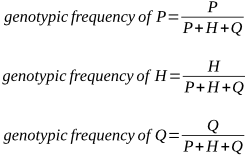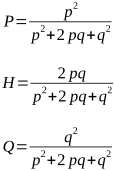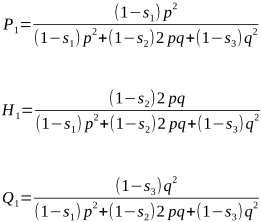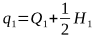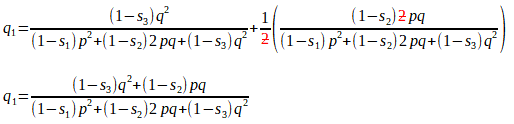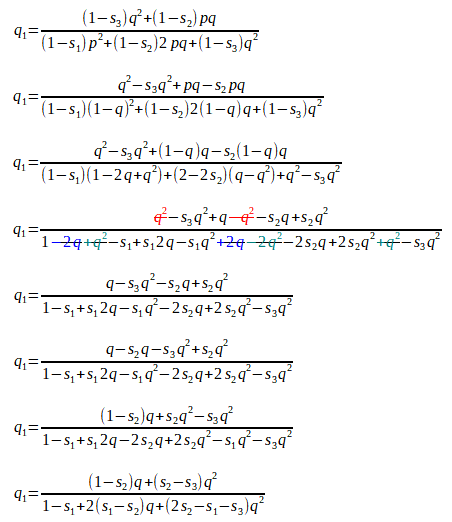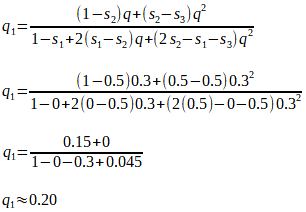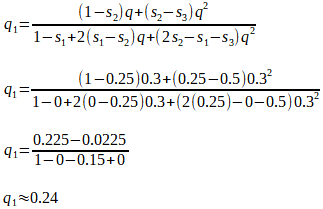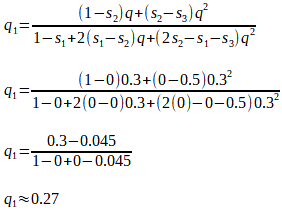This post revisits several concepts discussed in the Population Genetics category. I’ve linked to the relevant pages as they come up below if you need a refresher.
Assume a locus A with alleles A1 and A2. Either could be, but is not necessarily, dominant or recessive to the other.
p is the gene frequency of the A1 allele and q is the gene frequency of the A2 allele. (Similarly, p and q as used here do not necessarily ascribe dominance or recessiveness to either allele.)
P is the genotypic frequency of the A1A1 genotype.
H is the genotypic frequency of the A1A2 genotype.
Q is the genotypic frequency of the A2A2 genotype.
Remember that P + H + Q = 1. If there are three genotypes in a population, the proportion of each as a percentage (its frequency) must add up to 100%, or 1.
We can express the proportions of P, H and Q as:
Assume these frequencies occur in a population from which parents have not yet been selected to produce the next generation.
If their parents were randomly mated, then this population should be in a Hardy-Weinberg equilibrium, thus:
P = p2
H = 2pq
Q = q2
Substituting these values into the formulae above, we get:
But we need to take into account the degree of dominance with respect to fitness for each of the genotypes A1A1, A1A2 and A2A2.
Let s1 be the relative fitness difference for A1A1.
Let s2 be the relative fitness difference for A1A2.
Let s3 be the relative fitness difference for A2A2.
If A1A1 is the fittest genotype (produces the most offspring), then its fitness difference, relative to itself, is s1 = 0, and its relative fitness is (1 - s1) = (1 - 0) = 1.
If, hypothetically, the A1A2 genotype produces 25% fewer offspring than the A1A1 genotype, then the fitness difference, relative to the fittest genotype A1A1, is s2 = 0.25, and the relative fitness is (1 - s2) = (1 - 0.25) = 0.75. We can say that the A1A2 genotype is 75%, or 0.75 as fit as the A1A1 genotype.
More generally, we can say that:
the relative fitness value for A1A1 is (1 - s1),
the relative fitness value for A1A2 is (1 - s2), and
the relative fitness value for A2A2 is (1 - s3).
In other words:
the relative fitness of A1A1 is (1 - s1) of its genotypic frequency P, or
(1 - s1) × P = (1 - s1) × p2 = (1 - s1) p2
Similarly, we can state the relative fitness of A1A2 as (1 - s2)2pq, and the relative fitness of A2A2 as (1 - s3)q2.
We wish to select animals from this population to be the parents of the next generation. From this, their progeny become the next parents, with genotypic frequencies of P1, H1 and Q1. Can you see how the following formulae are the same as above, but this time we have taken into consideration the relative fitness values for each of P1, H1 and Q1:
In The Effect of Mating Systems on Gene and Genotypic Frequencies: Outbreeding, we saw how the gene frequency q = Q + ½H.
From this, the frequency of the A2 allele after selection is:
Substituting for Q1 and H1, we get:
[Here we have ’simply’ added two (elaborate) fractions with a common denominator. This is just a/c + b/c = a + b/c on steroids!]
As there are just two alleles A1 and A2, with the respective gene frequencies p and q , these must add up to one, as p + q = 1. We can rewrite this as p = 1 - q.
We can substitute (1 - q) for p, cancel some terms, and rearrange the rest to get:
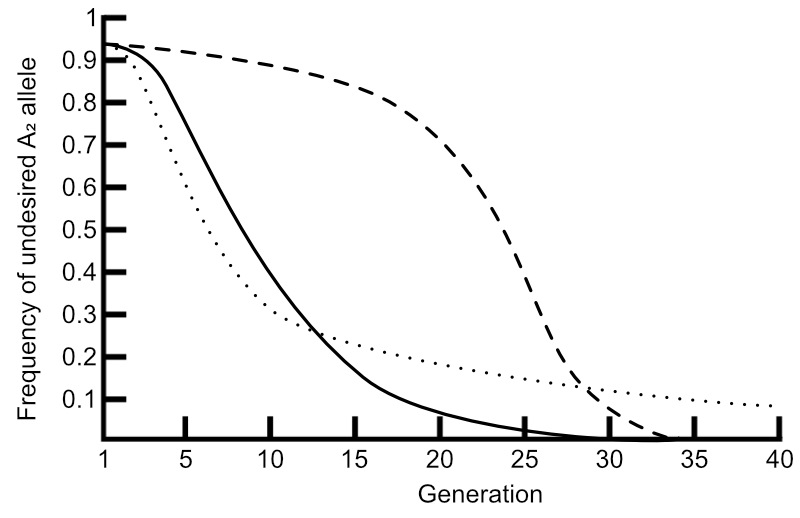
We now have a formula with which we can calculate the new gene frequency in the next population, given the initial gene frequency, the degree of dominance (if any), and the difference in fitness values (if any) of the various genotypes.
Next week we’ll run through some scenarios to see this in practice!