The Maths of Matings, Part 1
Having gone over confidence levels, it’s time to apply that and step through some maths!
Let’s now calculate confidence levels and the required number of test matings to be statistically confident that a tested animal is not a carrier of a recessive allele.
Everything below assumes that the tested animal is a sire, that there is one offspring from one mating, and that all mates (dams) are of the same type for the allele of interest. That is, they are either all known carriers, or are all daughters of the tested sire, or are all randomly selected from the same population.
Now let:
n = number of matings that produce an offspring
P[Dn] = probability (P) of detection (D) that at least one homozygous recessive offspring is born for n number of matings. This is our level of confidence in the test.
PAA = probability that a mate is homozygous dominant (’AA’) at the locus being tested.
PAa = probability that a mate is heterozygous (’Aa’) at the locus being tested.
Paa = probability that a mate is homozygous recessive (’aa’) at the locus being tested.
From all this:
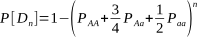
OK, so where did this come from, you may well be asking?! Let’s break this down bit by bit.
P[Dn], as stated above, is our level of confidence.
From last week, our level of confidence can also be written as 1 - α.
Thus P[Dn] = 1 - α.
Substitute α (alpha) with ![]() , and you have 1 -
, and you have 1 - ![]()
But what is that sum inside the parentheses?
Firstly, does it make sense to you that the (probability of homozygous recessive born) + (probability of no homozygous recessive born) must equal 1, or 100%?
In other words, that it is 100% certain that all offspring born must have some combination of alleles, regardless of what that combination is?
And from that, does it also make sense that to calculate the probability of at least one homozygous recessive offspring being born, that you could calculate the probabilities of no homozygous recessive offspring being born, and subtract that from 1 (100%)? We could write that as:
(probability of homozygous recessive born) = 1 - (probability of no homozygous recessive born)
From this, can you see how ![]() is the probability of no homozygous recessive born?
is the probability of no homozygous recessive born?
Going further:
PAA is the probability that a dam is homozygous dominant (’AA’) at the locus being tested. This animal by definition can only contribute ‘A’ alleles.
The probability that she will birth a non-recessive offspring is 1 (100%), whether or not the sire is a carrier.
The PAA is thus written as if it were 1 × PAA.
PAa is the probability that a dam is heterozygous (’Aa’) at the locus being tested.
If both the sire and female are carriers, there are three chances in four that a dominant phenotype will be born, hence we write 3/4PAa.
Paa is the probability that a dam is homozygous recessive (’aa’) at the locus being tested.
If the sire is a carrier, and the female is homozygous recessive, there are two chances in four that a dominant phenotype will be born, hence we write 1/2Paa.
To calculate the number of matings to give us enough confidence that a sire is not a carrier, we need to know (mating outcome 1) and (mating outcome 2), and so on for some number n such that the confidence level is high.
(mating outcome 1) and (mating outcome 2) and (mating outcome 3) and so on can be rewritten
(mating outcome 1) × (mating outcome 2) × (mating outcome 3) × (…), as in statistics, when we say ‘and’ we write ‘×’.
And (mating outcome 1) × (mating outcome 2) × (mating outcome 3) × (…) is the same as writing
(mating outcome)n, where n is the number of matings.
Hopefully ![]() isn’t so scarey-looking now?
isn’t so scarey-looking now?
Time to use it and work with some numbers!
Assume we have five known carriers to test a sire with. This means the probabilities of PAA and Paa must both be 0, and that of PAa is 1.
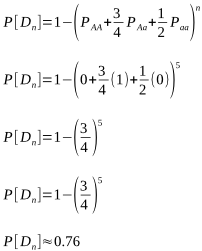
That isn’t a very high level of confidence, but let’s take an extreme and assume 30 known carriers:
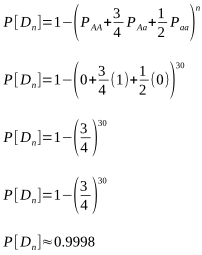
Now that is a very high level of confidence that the sire is not a carrier! But it would appear we could use a smaller number of females for almost as good a result. That would certainly be a more preferable use of dams. Rather than just punching in numbers at random to find the most favourable values for n and [Dn], the most practical approach is to solve for n outright.
How are you with logs?! (I don’t want to dwell on logs too much here but I have written a little supplementary post here to explain how the formula was rearranged below.)
Let’s rearrange to solve for n:
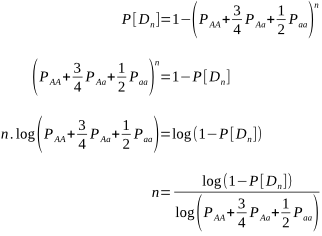
The number of matings required to give us a 95% level of confidence that no homozygous recessives will be born, and that the sire is not a carrier, using known carrier dams is:
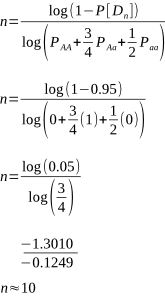
This is an interesting example to make a point with. The solution for n, to two decimal places, was actually 10.42. Hence the rounding down to 10. But it’s so close to 10.5 that maybe it should be rounded up to 11 for an extra margin of safety.
Indeed, you should always factor in additional matings anyway, to account for matings that don’t take or aborted foetuses. The calculated number of matings assumes a live birth for every mating, ie it assumes n number of successful matings.
The above assumed one offspring per mating and that all mates were of the same type. We’ll stop there as that was quite a lot of information to absorb! Next week we’ll go over the maths for other scenarios involving multiple groups of mates and multiple births per mate.
Leave a comment